Nye publikasjoner
Nye genetiske mekanismer kan være et terapeutisk mål mot gliom
Sist anmeldt: 02.07.2025

Alt iLive-innhold blir gjennomgått med medisin eller faktisk kontrollert for å sikre så mye faktuell nøyaktighet som mulig.
Vi har strenge retningslinjer for innkjøp og kun kobling til anerkjente medieområder, akademiske forskningsinstitusjoner og, når det er mulig, medisinsk peer-evaluerte studier. Merk at tallene i parenteser ([1], [2], etc.) er klikkbare koblinger til disse studiene.
Hvis du føler at noe av innholdet vårt er unøyaktig, utdatert eller ellers tvilsomt, velg det og trykk Ctrl + Enter.
En studie fra laboratoriet til Shi-Yuan Cheng, PhD, professor ved Ken og Ruth Daveys avdeling for nevro-onkologi ved nevrologisk avdeling, har identifisert nye mekanismer som ligger til grunn for alternative RNA-spleisingshendelser i gliomtumorceller som kan tjene som nye terapeutiske mål. Studieresultatene er publisert i Journal of Clinical Investigation.
«Vi har funnet en annen måte å behandle gliom gjennom linsen til alternativ spleising og oppdaget nye mål som ikke har blitt identifisert tidligere, men som er viktige for gliommalignitet», sa Xiao Song, MD, PhD, førsteamanuensis i nevrologi og hovedforfatter av studien.
Gliomer er den vanligste typen primær hjernesvulst hos voksne og stammer fra gliaceller, som befinner seg i sentralnervesystemet og støtter nærliggende nevroner. Gliomer er svært resistente mot standardbehandlinger, inkludert stråling og cellegift, på grunn av svulstens genetiske og epigenetiske heterogenitet, noe som understreker behovet for å finne nye terapeutiske mål.
Tidligere forskning fra Cheng-laboratoriet, publisert i tidsskriftet Cancer Research, viste at den viktige spleisingsfaktoren SRSF3 er betydelig forhøyet i gliomer sammenlignet med normale hjerner, og SRSF3-regulert RNA-spleising fremmer gliomvekst og -progresjon ved å påvirke flere cellulære prosesser i tumorceller.
RNA-spleising er en prosess som involverer fjerning av introner (ikke-kodende regioner av RNA) og sammenføyning av eksoner (kodende regioner) for å danne et modent mRNA-molekyl som støtter genuttrykk i en celle.
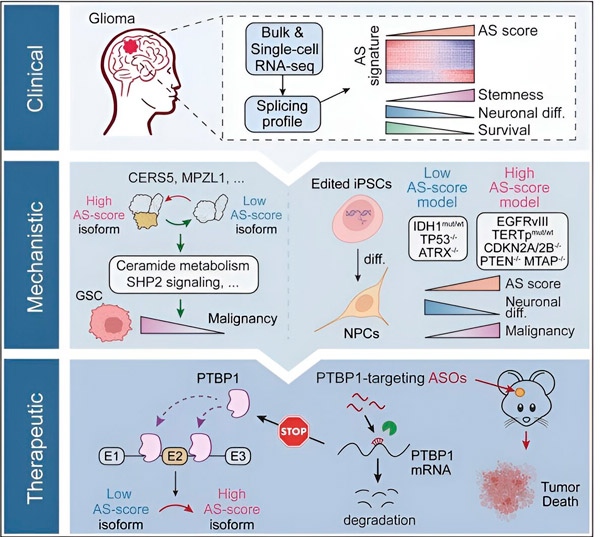
I den foreliggende studien hadde forskerne som mål å identifisere endringer i alternativ spleising i gliomtumorceller, mekanismene som ligger til grunn for disse endringene, og bestemme deres potensial som terapeutiske mål.
Ved hjelp av beregningsmetoder og RNA-sekvenseringsteknologier undersøkte forskerne spleisingendringer i gliomtumorceller fra pasientprøver. For å bekrefte disse endringene brukte de CRISPR-genredigeringsteknologier for å introdusere forskjellige gliomdrivermutasjoner i gliommodeller avledet fra humant induserte pluripotente stamceller (iPSC-er).
De fant at disse spleisingsendringene forsterkes av en variant av epidermal vekstfaktorreseptor III (EGFRIII), som er kjent for å være overuttrykt i mange svulster, inkludert gliomer, og hemmes av en mutasjon i IDH1-genet.
Forskere har bekreftet funksjonen til to RNA-spleisingshendelser som skaper forskjellige proteinisoformer med forskjellige aminosyresekvenser.
"Bare én av disse isoformene kan fremme tumorvekst, i motsetning til den andre isoformen, som normalt uttrykkes i den normale hjernen. Tumorer utnytter denne mekanismen til å selektivt uttrykke den tumorfremmende isoformen over den normale hjerneisoformen," sa Song.
Teamet analyserte deretter oppstrøms RNA-bindende proteiner og fant ut at PTBP1-genet regulerer tumorfremmende RNA-spleising i gliomceller. Ved å bruke en ortotopisk gliommodell hos immunsviktige mus, målrettet forskerne PTBP1 med antisense-oligonukleotid (ASO)-basert terapi, som til slutt undertrykte tumorvekst.
«Dataene våre fremhever rollen til alternativ RNA-spleising i påvirkning av gliommalignitet og heterogenitet, og dens potensial som en terapeutisk sårbarhet for behandling av voksne gliomer», skrev forfatterne av studien.
Det neste steget for forskerne er å utforske potensialet for å målrette PTBP1 for å fremkalle en antitumorimmunrespons, sa Song.
"Ved å bruke long-read RNA-sekvenseringsanalyse fant vi at målretting av PTBP1 i gliomaceller resulterer i produksjon av flere alternativt spleisete transkripter som er fraværende i normalt vev. Så vårt neste prosjekt er å finne ut om denne isoformen kan generere noen antigener slik at immunforsvaret bedre kan gjenkjenne svulsten," sa Song.
Song la også til at teamet deres er interessert i å analysere spleisingendringer i ikke-tumorceller fra gliompasienter, som immunceller.
«Vi vet allerede at spleising er svært viktig for å regulere funksjonen i cellen, så den bør ikke bare regulere svulstmalignitet, men den kan også regulere funksjonen til immunceller for å avgjøre om de effektivt kan drepe kreft. Så vi gjør også noen bioinformatiske analyser i tumorinfiltrerte immunceller for å se om det er en endring i spleising etter at en immuncelle har infiltrert svulsten.»
"Målet vårt er å bestemme rollen til alternativ spleising i utformingen av det immunsuppressive svulstmikromiljøet og identifisere potensielle mål for å forbedre effekten av immunterapier i gliom," sa Song.
